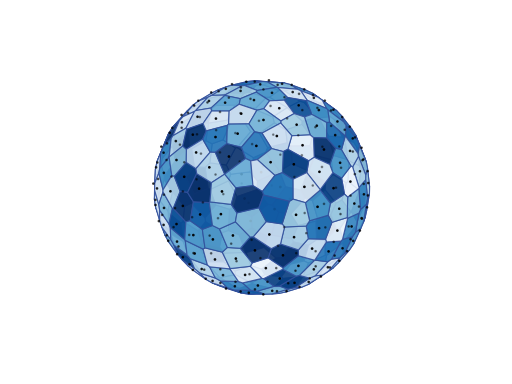
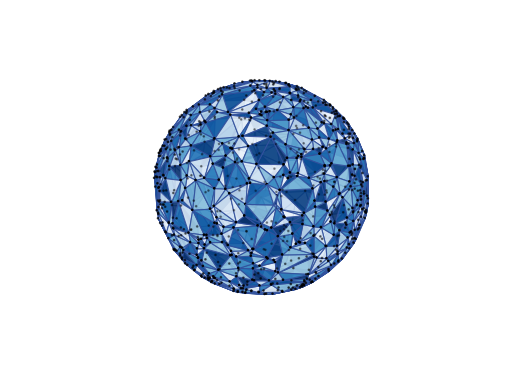
Spherical Meshes¶
Module: pycsphere.mesh
Routines for deterministic and random spherical point sets.
SphericalPointSet(resolution[, separation, …])Base class for spherical point sets.
FibonacciPointSet(N[, lonlat])Fibonacci point set.
HEALPixPointSet(nside[, lonlat, nest])HEALPix point set.
RandomPointSet(N[, seed, lonlat])Random uniform point set.
-
class
SphericalPointSet(resolution: int, separation: Optional[float] = None, nodal_width: Optional[float] = None, lonlat: bool = False)[source]¶ Bases:
abc.ABCBase class for spherical point sets.
Any subclass/instance of this class must implement the abstract method
generate_point_setwhich returns the point set cartesian/spherical coordinates.A spherical point set is a collection \(\Theta_N=\{\mathbf{r}_1, \ldots, \mathbf{r}_N\}\subset \mathbb{S}^2\) of unit vectors (directions).
Notes
The class
SphericalPointSetpossesses a few class/object attributes which allow to describe the properties of the point set. The terminology is the same as [PointSets]:A sequence of point sets \(\{\Theta_N\}_{n\in\mathbb{N}}\subset \mathcal{P}(\mathbb{S}^2)\) is called
equidistributedif the sequence of normalised atomic measures \(\nu_N(A):=|A\cap \Theta_N|/N\) converges w.r.t. the weak*-topology towards the Lebesgue measure as \(N\) grows to infinity.The separation of a point set is defined as \(\delta(\Theta_N):=\min_{\mathbf{r}\neq \mathbf{s}\in\Theta_N} \|\mathbf{r}- \mathbf{s}\|_2\). A set is said
well_separatedif \(\delta(\Theta_N)\geq c N^{-1/2}\) for someseparation_cstconstant \(c>0\) and \(N\geq 2\).The
nodal_widthof a point set is defined as \(\eta(\Theta_N):=\max_{\mathbf{s}\in\mathbb{S}^2}\min_{\mathbf{r}\in\Theta_N} \|\mathbf{r}- \mathbf{s}\|_2\). A set is said to havegood_coveringif \(\eta(\Theta_N)\leq C N^{-1/2}\) for somenodal_width_cst\(C>0\) and \(N\geq 2\).The
mesh_ratioof a point set is the ratio between its nodal width and separation. If the mesh ratio is asymptotically bounded, then the point set is saidquasi_uniform. A lower bound on the mesh ratio isminimal_mesh_ratio = (np.sqrt(5) - 1) / 2.
-
classmethod
arc2angle(arc_length: Union[numpy.ndarray, float], deg: bool = False) → Union[numpy.ndarray, float][source]¶ Convert an arc length to an angular section.
-
classmethod
angle2arc(angle: Union[numpy.ndarray, float], deg: bool = False) → Union[numpy.ndarray, float][source]¶ Compute the arc length of a given angular section.
-
classmethod
angle2res(angular_resolution: float, deg: bool = True) → int[source]¶ Compute the minimal point set size required to achieve a desired (angular) nodal width.
-
__init__(resolution: int, separation: Optional[float] = None, nodal_width: Optional[float] = None, lonlat: bool = False)[source]¶ - Parameters
resolution (int) – Resolution (size) of the point set.
separation (Optional[float]) – Separation of the point set (
Noneif unknown).nodal_width (Optional[float]) – Nodal width of the point set (
Noneif unknown).lonlat (bool) – If
True, longitude/latitude spherical coordinates (in degrees) are used. Otherwise, longitude/co-latitude (in radians) are used.
-
abstract
generate_point_set() → Tuple[numpy.ndarray, numpy.ndarray][source]¶ Generate the cartesian/spherical coordinates of the point set.
- Returns
Cartesian/spherical coordinates of the point set. The latter are also stored in the attributes
self.vecandself.dirrespectively. The attributeself.vechas shape(3, N)and contains the cartesian coordinates of the points in the set. The attributeself.dirhas shape(2, N)and contains the spherical coordinates of the points in the set:If
self.latlon==True, longitudes and latitudes in degrees,If
self.latlon==False, longitudes and co-latitudes in radians.
- Return type
Tuple[np.ndarray, np.ndarray]
-
plot_tessellation(facecmap: Optional[str] = 'Blues_r', ncols: int = 20, edgecolor: str = '#32519D', cell_centers: bool = True, markercolor: str = 'k', markersize: float = 2, linewidths=1, alpha: float = 0.9, seed: int = 0)[source]¶ Plots the tessellation defined by the point set.
- Parameters
facecmap (Optional[str]) – Color map to be used for random coloring the tessellation faces.
ncols (int) – Number of colors in the color map.
edgecolor (str) – Color of the cell edges.
cell_centers (bool) – Whether or not the cell centers are displayed.
markercolor (str) – Color of the cell centers.
markersize (float) – Size of the cell centers.
linewidths (float) – Width of the cell edges.
alpha (float) – Transparency of the cell faces.
seed (int) – Seed for random coloring reproducibility.
Warning
This method can be very slow for large point sets (>10’000 points)!
-
plot_delaunay_cells(facecmap: Optional[str] = 'Blues_r', ncols: int = 20, edgecolor: str = '#32519D', cell_centers: bool = True, markercolor: str = 'k', markersize: float = 2, linewidths=1, alpha: float = 0.9, seed: int = 0)[source]¶ Plots the tessellation defined by the set’s Delaunay triangulation.
- Parameters
facecmap (Optional[str]) – Color map to be used for random coloring the tessellation faces.
ncols (int) – Number of colors in the color map.
edgecolor (str) – Color of the cell edges.
cell_centers (bool) – Whether or not the cell centers are displayed.
markercolor (str) – Color of the cell centers.
markersize (float) – Size of the cell centers.
linewidths (float) – Width of the cell edges.
alpha (float) – Transparency of the cell faces.
seed (int) – Seed for random coloring reproducibility.
Warning
This method can be very slow for large point sets (>10’000 points)!
-
plot_voronoi_cells(facecmap: Optional[str] = 'Blues_r', ncols: int = 20, edgecolor: str = '#32519D', cell_centers: bool = True, markercolor: str = 'k', markersize: float = 2, linewidths=1, alpha: float = 0.9, seed: int = 0)[source]¶ Plots the tessellation defined by the set’s Voronoi decomposition.
- Parameters
facecmap (Optional[str]) – Color map to be used for random coloring the tessellation faces.
ncols (int) – Number of colors in the color map.
edgecolor (str) – Color of the cell edges.
cell_centers (bool) – Whether or not the cell centers are displayed.
markercolor (str) – Color of the cell centers.
markersize (float) – Size of the cell centers.
linewidths (float) – Width of the cell edges.
alpha (float) – Transparency of the cell faces.
seed (int) – Seed for random coloring reproducibility.
Warning
This method can be very slow for large point sets (>10’000 points)!
-
property
angular_nodal_width¶ Angular nodal width.
-
class
FibonacciPointSet(N: int, lonlat: bool = False)[source]¶ Bases:
pycsphere.mesh.SphericalPointSetFibonacci point set.
Examples
from pycsphere.mesh import FibonacciPointSet N = FibonacciPointSet.angle2N(angular_resolution=10) fib = FibonacciPointSet(N, lonlat=True) fib.plot_delaunay_cells() fib.plot_tessellation()
Notes
Points in the Fibonacci point set are arranged uniformly along a spiral pattern on the sphere linking the two poles (see Section 2 of [PointSets] for a definition of the point set). The Fibonacci point set is defined for odd resolutions \(2N+1\) only. It is well-separated, has good covering and is hence quasi-uniform. It is also equidistributed. Its Voronoi cells are however irregular and do not have the same shape/area. The points are not arranged on isolatitude tracks. Finally it is non hierarchical (Fibonacci point sets at two different resolutions are not contained in one another).
Todo
Add support for local Fibonacci meshes.
-
classmethod
angle2N(angular_resolution: float, deg: bool = True) → int[source]¶ Minimal parameter \(N\) to achieve a prescribed angular nodal width.
-
__init__(N: int, lonlat: bool = False)[source]¶ - Parameters
N (int) – Defines the point set resolution \(2N+1\).
lonlat (bool) – Convention for the spherical coordinates (see help of
__init__()).
-
generate_point_set() → Tuple[numpy.ndarray, numpy.ndarray][source]¶ Generate the cartesian/spherical coordinates of the point set.
- Returns
Cartesian/spherical coordinates of the point set. The latter are also stored in the attributes
self.vecandself.dirrespectively. The attributeself.vechas shape(3, N)and contains the cartesian coordinates of the points in the set. The attributeself.dirhas shape(2, N)and contains the spherical coordinates of the points in the set:If
self.latlon==True, longitudes and latitudes in degrees,If
self.latlon==False, longitudes and co-latitudes in radians.
- Return type
Tuple[np.ndarray, np.ndarray]
-
classmethod
-
class
HEALPixPointSet(nside: int, lonlat: bool = False, nest=False)[source]¶ Bases:
pycsphere.mesh.SphericalPointSetHEALPix point set.
Examples
from pycsphere.mesh import HEALPixPointSet nside = HEALPixPointSet.angle2nside(angular_resolution=10) healpix = HEALPixPointSet(nside, lonlat=True) healpix.plot_delaunay_cells() healpix.plot_voronoi_cells() healpix.plot_tessellation()
Notes
Developed by NASA for fast data analysis of the cosmic microwave background (CMB), the Hierarchical Equal Area iso-Latitude Pixelization (HEALPix) was designed to have three properties essential for computational efficiency in discretising functions on \(\mathbb{S}^2\):
The sphere is hierarchically tessellated into curvilinear quadrilaterals.
The pixelisation is an equal area partition of \(\mathbb{S}^2\).
The point sets are distributed along iso-latitude lines.
A formal definition of the point set is provided Section 2 of [PointSets]. We use here the
healpypackage. The HEALPix point set is defined for resolutions \(N=12k^2\) only (parameter \(k\) is callednside). It is well-separated, has good covering and is hence quasi-uniform. It is also equidistributed. Its tessellation cells are moreover regular and have the same shape/area. Finally it is hierarchical (point sets at two different resolutions are nested in one another).Todo
Add spatial filtering routines for local HEALPix meshes.
-
classmethod
angle2nside(angular_resolution: float, deg: bool = True) → int[source]¶ Minimal parameter \(k\) (
nside) to achieve a prescribed angular nodal width.
-
__init__(nside: int, lonlat: bool = False, nest=False)[source]¶ - Parameters
nside (int) – Defines the point set resolution
12 * nside ** 2.nsidemust be a power of two.lonlat (bool) – Convention for the spherical coordinates (see help of
__init__()).nest (bool) – If
True, the HEALPix mesh is stored in NESTED ordering (efficient for pixel querying routines). IfFalse, the HEALPix mesh is stored in RING ordering (efficient for spherical harmonics transforms).
-
generate_point_set() → Tuple[numpy.ndarray, numpy.ndarray][source]¶ Generate the cartesian/spherical coordinates of the point set.
- Returns
Cartesian/spherical coordinates of the point set. The latter are also stored in the attributes
self.vecandself.dirrespectively. The attributeself.vechas shape(3, N)and contains the cartesian coordinates of the points in the set. The attributeself.dirhas shape(2, N)and contains the spherical coordinates of the points in the set:If
self.latlon==True, longitudes and latitudes in degrees,If
self.latlon==False, longitudes and co-latitudes in radians.
- Return type
Tuple[np.ndarray, np.ndarray]
-
plot_tessellation(facecmap: Optional[str] = 'Blues_r', ncols: int = 20, edgecolor: str = '#32519D', cell_centers: bool = True, markercolor: str = 'k', markersize: float = 2, linewidths=1, alpha: float = 0.9, seed: int = 0)[source]¶ Plots the tessellation defined by the point set.
- Parameters
facecmap (Optional[str]) – Color map to be used for random coloring the tessellation faces.
ncols (int) – Number of colors in the color map.
edgecolor (str) – Color of the cell edges.
cell_centers (bool) – Whether or not the cell centers are displayed.
markercolor (str) – Color of the cell centers.
markersize (float) – Size of the cell centers.
linewidths (float) – Width of the cell edges.
alpha (float) – Transparency of the cell faces.
seed (int) – Seed for random coloring reproducibility.
Warning
This method can be very slow for large point sets (>10’000 points)!
-
class
RandomPointSet(N: int, seed: int = 0, lonlat: bool = False)[source]¶ Bases:
pycsphere.mesh.SphericalPointSetRandom uniform point set.
Examples
from pycsphere.mesh import RandomPointSet res = RandomPointSet.angle2res(angular_resolution=10) rnd = RandomPointSet(res, lonlat=True) rnd.plot_delaunay_cells() rnd.plot_tessellation()
Notes
Properties of spherical random point sets are discussed in Section 2 of [PointSets]. They are equidistributed but not quasi-uniform, well-separated or with good-coverage. They are also of course not hierarchical and yield highly irregular tessellation cells with different shapes and areas.
-
classmethod
angle2res(angular_resolution: float, deg: bool = True) → int[source]¶ Compute the minimal point set size required to achieve a desired (angular) nodal width.
-
__init__(N: int, seed: int = 0, lonlat: bool = False)[source]¶ - Parameters
N (int) – Resolution of the random point set.
seed (int) – Seed for reproducibility of the random point set.
lonlat (bool) – Convention for the spherical coordinates (see help of
__init__()).
-
generate_point_set() → Tuple[numpy.ndarray, numpy.ndarray][source]¶ Generate the cartesian/spherical coordinates of the point set.
- Returns
Cartesian/spherical coordinates of the point set. The latter are also stored in the attributes
self.vecandself.dirrespectively. The attributeself.vechas shape(3, N)and contains the cartesian coordinates of the points in the set. The attributeself.dirhas shape(2, N)and contains the spherical coordinates of the points in the set:If
self.latlon==True, longitudes and latitudes in degrees,If
self.latlon==False, longitudes and co-latitudes in radians.
- Return type
Tuple[np.ndarray, np.ndarray]
-
classmethod