Solving Inverse Problems with Pycsou¶
Pycsou is a Python 3 package for solving linear inverse problems
with state-of-the-art proximal algorithms. The library provides abstract
classes for the main building blocks of generic penalised convex
optimisation problems:
linear operators,
loss functionals,
penalty functionals,
proximal algorithms.
Penalised convex optimisation problems can then be constructed by adding, subtracting, scaling, composing, exponentiating or stacking together the various linear operators and functionals shipped with Pycsou (or custom, user-defined ones).
Multidimensional Maps¶
Pycsou’s base classes for functionals and linear operators both inherit
from the abstract class Map with signature:
class Map(shape: Tuple[int, int], is_linear: bool = False,
is_differentiable: bool = False):
This is the base class for multidimensional maps (potentially nonlinear) \(\mathbf{h}:\mathbb{R}^M\to\mathbb{R}^N\).
Any instance/subclass of this class must at least implement the abstract
method __call__ for pointwise evaluation.
Operations on Map Instances¶
The class Map supports the following arithmetic operators +,
-, *, @, ** and /, implemented with the class
methods __add__/__radd__, __sub__/__neg__, __mul__/__rmul__,
__matmul__, __pow__,__truediv__. Such arithmetic operators
can be used to add, substract, scale, compose, exponentiate or
evaluate Map instances. The __call__ methods of Map objects
constructed this way are automatically computed:
>>> f3 = f1 / 3 + np.pi * f2
>>> np.allclose(f3(x), f1(x) / 3 + np.pi * f2(x))
True
>>> h3 = h1 * 3 - (h2 ** 2) / 6
>>> np.allclose(h3(x), h1(x) * 3 - (h2(h2(x))) / 6)
True
Note that multiplying a map with an array is the same as evaluating the map at the array:
>>> np.allclose(h * x, h(x))
True
The multiplication operator @ can also be used in place of *, in
compliance with Numpy’s interface:
>>> np.allclose(h * x, h @ x), np.allclose((h1 * h2)(x), (h1 @ h2)(x))
True, True
Finally, maps can be shifted via the method shifter:
>>> h=g.shifter(shift=2 * x)
>>> np.allclose(h(x), g(x + 2 * x))
True
Differentiable Maps¶
An important subclass of Map is DifferentiableMap which is the
base class for differentiable maps. It has the following signature:
class DifferentiableMap(shape: Tuple[int, int],is_linear: bool = False,
lipschitz_cst: float = inf,
diff_lipschitz_cst: float = inf):
Any instance/subclass of this class must implement the abstract methods
__call__ and jacobianT which returns the transpose of the
Jacobian matrix
of the multidimensional \(\mathbf{h}=[h_1, \ldots, h_N]: \mathbb{R}^M\to\mathbb{R}^N\) at a given point \(\mathbf{x}\in\mathbb{R}^M\).
Operations on DifferentiableMap Instances¶
Standard arithmetic operators can also be used on DifferentiableMap
instances so as to add, substract, scale, compose, exponentiate or
evaluate them. The attributes lipschitz_cst, diff_lipschitz_cst
and the method jacobianT are automatically updated using standard
differentiation rules.
>>> map_ = f * g
>>> np.allclose(map_.lipschitz_cst, f.lipschitz_cst * g.lipschitz_cst)
True
>>> np.allclose(map_.jacobianT(x), g.jacobianT(x) * f.jacobianT(g(x)))
True
Functionals¶
Functionals are (real) single-valued maps
\(h:\mathbb{R}^N\to \mathbb{R}\). They can be implemented via a
subclass of Map called Functional:
class Functional(dim: int,
data: Optional[numpy.ndarray] = None,
is_differentiable: bool = False,
is_linear: bool = False)
For differentiable functionals, the subclass
DifferentiableFunctional can be used. The latter admits a method
gradient which is an alias for the abstract method jacobianT.
Note
Reminder: for a functional \(h\), \(\mathbf{J}^T_h(\mathbf{x})=\nabla h (\mathbf{x})\).
Proximable Functionals¶
We say that a functional \(f:\mathbb{R}^N\to \mathbb{R}\) is proximable is its proximity operator
admits a simple closed-form expression or can be evaluated efficiently and with high accuracy.
They are represented by the subclass ProximableFunctional. The
latter has signature:
class ProximableFunctional(dim: int,
data: Optional[numpy.ndarray] = None,
is_differentiable: bool = False,
is_linear: bool = False)
Every subclass/instance of ProximableFunctional must at least
implement the abstract methods __call__ and prox.
Note
See Cost Functionals for a list of common functionals already implemented in Pycsou.
Operations on Proximable Functionals¶
For the following basic operations, the proximal operator can be automatically updated:
Postcomposition: \(g(\mathbf{x})=\alpha f(\mathbf{x})\),
Precomposition: \(g(\mathbf{x})= f(\alpha\mathbf{x}+b)\) or \(g(\mathbf{x})= f(U\mathbf{x})\) with \(U\) a unitary operator,
Affine Sum: \(g(\mathbf{x})= f(\mathbf{x})+\mathbf{a}^T\mathbf{x}.\)
>>> from pycsou.func.penalty import L1Norm
>>> func = L1Norm(dim=10)
>>> x = np.arange(10); tau=0.1
>>> np.allclose((2 * func).prox(x, tau), func.prox(x, 2 * tau))
True
>>> np.allclose((func * 2).prox(x, tau), func.prox(x * 2, 4 * tau)/2)
True
>>> np.allclose(func.shifter(x/2).prox(x, tau), func.prox(x+x/2, tau)-x/2)
True
Horizontal Stacking of Proximable Functionals¶
The class ProxFuncHStack allows to stack many functionals
\(\{f_i:\mathbb{R}^{N_i}\to \mathbb{R}, i=1,\ldots, k\}\)
horizontally:
The proximity operator of the stacked functional \(h\) is moreover computed automatically (and soon in parallel) via the formula:
Example: Implementing New Differentiable Functionals¶
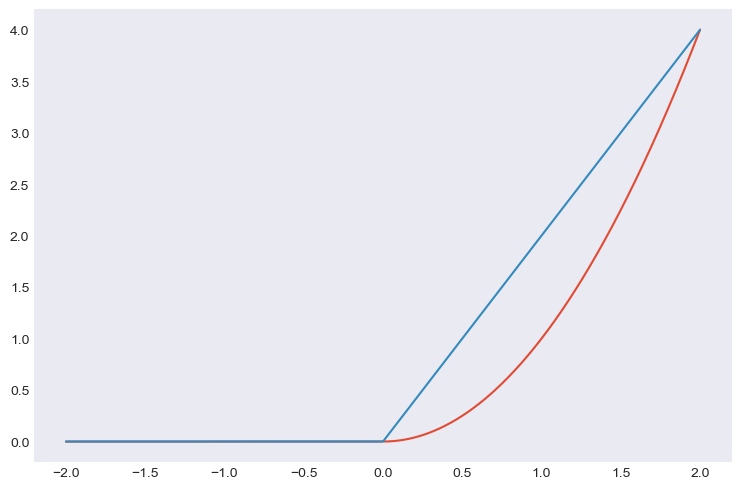
from pycsou.core import DifferentiableFunctional
class OneSidedSquaredL2Norm(DifferentiableFunctional):
def __init__(self, dim: int):
super(OneSidedSquaredL2Norm, self).__init__(dim=dim, diff_lipschitz_cst=2)
def __call__(self, x: np.ndarray) -> np.ndarray: #Implement abstract method __call__
return np.sum(x**2 * (x >= 0))
def jacobianT(self, x: np.ndarray) -> np.ndarray: #Implement abstract method jacobianT
return 2 * x * (x >= 0)
x=np.linspace(-2,2,1000)
func=OneSidedSquaredL2Norm(dim=1)
y = [func(t) for t in x]; dy = [func.gradient(t) for t in x]
plt.plot(x,y); plt.plot(x,dy)
Linear Operators¶
The base class for linear operators
\(\mathbf{L}:\mathbb{R}^N\to \mathbb{R}^M\) is LinearOperator, a
subclass of DifferentiableMap with signature:
class LinearOperator(shape: Tuple[int, int], dtype: Optional[type] = None,
is_explicit: bool = False, is_dense: bool = False,
is_sparse: bool = False, is_dask: bool = False,
is_symmetric: bool = False,
lipschitz_cst: float = inf)
Any instance/subclass of this class must at least implement the abstract
methods __call__ for forward evaluation
\(\mathbf{L}\mathbf{x}\) and adjoint for backward
evaluation \(\mathbf{L}^\ast\mathbf{y}\) where
\(\mathbf{L}^\ast:\mathbb{R}^M\to \mathbb{R}^N\) is the adjoint of
\(\mathbf{L}\) defined as:
Matrix-Free Operators¶
Pycsou’s linear operators are inherently matrix-free: the operator
\(\mathbf{L}\) needs not be stored as an array since the methods
__call__ (alias matvec) and adjoint can be used to perform
matrix-vector products \(\mathbf{L}\mathbf{x}\) and
\(\mathbf{L}^\ast\mathbf{y}\) respectively. This is particularly
useful when the dimensions \(N\) and \(M\) are very large
(e.g. in image processing) and \(\mathbf{L}\) cannot be stored in
memory as a Numpy array.
The class LinearOperator can be thought as an
interface-compatible overload of the standard matrix-free classes
pylops.LinearOperator and scipy.sparse.linalg.LinearOperator
from PyLops and Scipy respectively.
Pycsou’s LinearOperator introduces notably the method jacobianT,
useful for automatic differentiation when composing linear operators
with differentiable functionals:
It also introduces convenience linear algebra methods such as
eigenvals, svds, pinv, cond, etc.
Note
See Linear Operators for a list of matrix-free linear operators already implemented in Pycsou.
Explicit Operators¶
Sometimes it can be cumbersome to specify an operator in matrix-free
form. In which case, Pycsou’s class ExplicitLinearOperator can be
used to construct linear operators from array-like representations:
class ExplicitLinearOperator(array: Union[numpy.ndarray,
scipy.sparse.base.spmatrix,
dask.array.core.Array],
is_symmetric: bool = False)
This class takes as input Numpy arrays, Scipy sparse matrices (in any sparse format) or Dask distributed arrays.
Finally, matrix-free operators can be converted into explicit operators
via the methods todense or tosparse (useful for
visualisation/debugging but often memory intensive).
Operations on Linear Operators¶
Just like DifferentiableMap, the class LinearOperator supports
the whole set of arithmetic operations: +, -, *, @,
** and /. ’
The abstract methods __call__, jacobianT and adjoint of
LinearOperator instances resulting from arithmetic operations are
automatically updated.
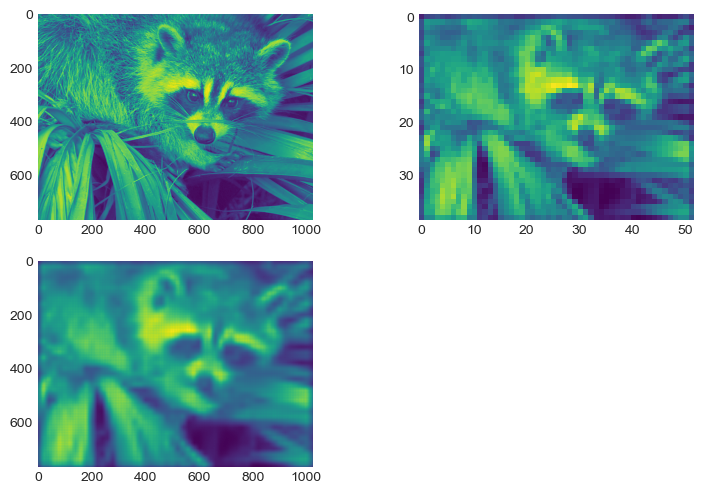
from pycsou.linop import Convolve2D, DownSampling
input_image = face(gray=True)
filter_size = 40
filter_support = np.linspace(-3, 3, filter_size)
gaussian_filter = np.exp(-(filter_support[:, None]
** 2 + filter_support[None, :] ** 2) / 2 * (0.5))
FilterOp = Convolve2D(size=input_image.size,
filter=gaussian_filter, shape=input_image.shape)
DownSamplingOp = DownSampling(
size=input_image.size, downsampling_factor=20, shape=input_image.shape)
BlurringOp = DownSamplingOp * FilterOp # Compose a filtering operator with a downsampling operator.
blurred_image = (BlurringOp * input_image.flatten()
).reshape(DownSamplingOp.output_shape) # Method __call__ of composite operator is available.
backproj_image = BlurringOp.adjoint(
blurred_image.flatten()).reshape(input_image.shape) # Method adjoint of composite operator is available.
plt.figure()
plt.subplot(2, 2, 1)
plt.imshow(input_image)
plt.subplot(2, 2, 2)
plt.imshow(blurred_image)
plt.subplot(2, 2, 3)
plt.imshow(backproj_image)
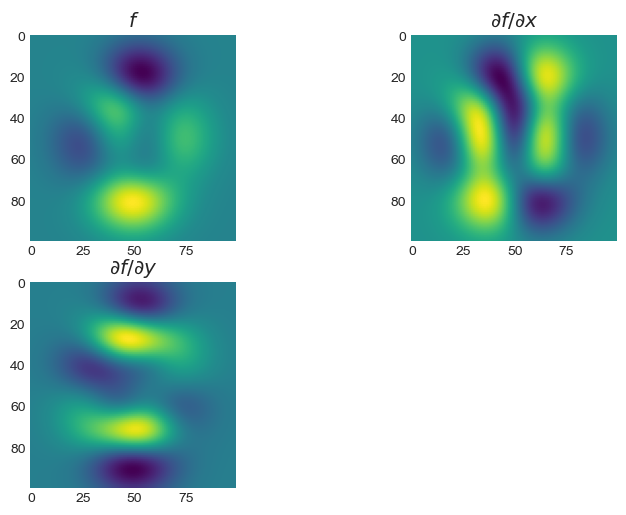
It is also possible to stack LinearOperator instances
horizontally/vertically via the class LinOpStack:
from pycsou.linop import FirstDerivative, LinOpStack
from pycsou.util import peaks
x = np.linspace(-2.5, 2.5, 100)
X,Y = np.meshgrid(x,x)
Z = peaks(X, Y)
D1 = FirstDerivative(size=Z.size, shape=Z.shape, axis=1,
kind='centered')
D2 = FirstDerivative(size=Z.size, shape=Z.shape, axis=0,
kind='centered')
Gradient = LinOpStack(D1, D2, axis=0) # Form the gradient by stacking 1D derivative operators
DZ=Gradient(Z.flatten())
plt.figure(); plt.subplot(2,2,1)
plt.imshow(Z)
plt.title('$f$'); plt.subplot(2,2,2)
plt.imshow(DZ[:Z.size].reshape(Z.shape))
plt.title('$\\partial f/\\partial x$'); plt.subplot(2,2,3)
plt.imshow(DZ[Z.size:].reshape(Z.shape))
plt.title('$\\partial f/\\partial y$')
Example: Implementing New Linear Operators¶
from pycsou.core import LinearOperator
class RepCol(LinearOperator):
def __init__(self, size: int, reps: int, dtype: type = np.float64):
self.reps = reps
super(RepCol, self).__init__(shape=(size*reps, size))
def __call__(self, x: np.ndarray) -> np.ndarray:
return np.tile(x[:,None], (1, self.reps)).flatten()
def adjoint(self, y: np.ndarray) -> np.ndarray:
return np.sum(y.reshape(self.shape[1], reps), axis=-1).flatten()
x =np.arange(4)
Op=RepCol(x.size, 5)
y=Op(x).reshape(x.size,5)
print(y)
[[0 0 0 0 0]
[1 1 1 1 1]
[2 2 2 2 2]
[3 3 3 3 3]]
Pycsou Class Diagram¶
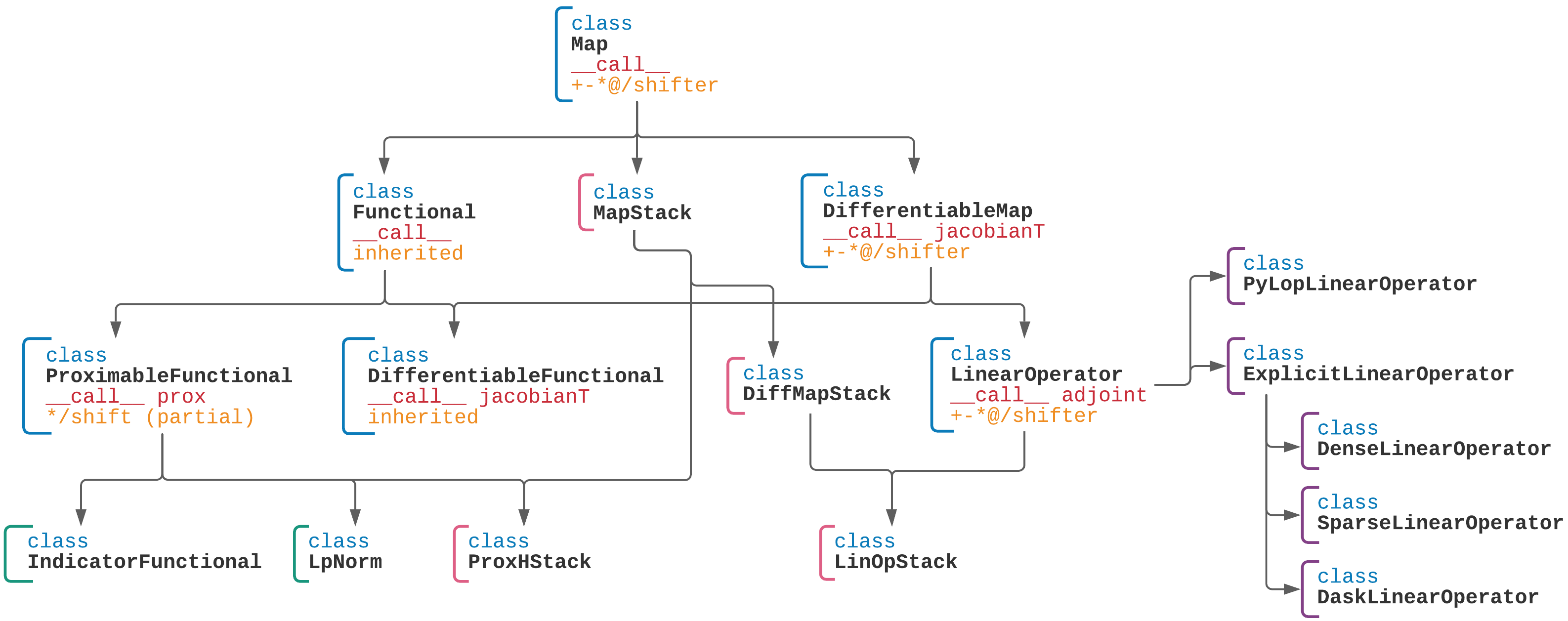
Algorithms¶
The base class for Pycsou’s iterative algorithms is
GenericIterativeAlgorithm:
class GenericIterativeAlgorithm(objective_functional: pycsou.core.map.Map,
init_iterand: Any,
max_iter: int = 500,
min_iter: int = 10,
accuracy_threshold: float = 0.001,
verbose: Optional[int] = None)
Any instance/subclass of this class must at least implement the abstract
methods update_iterand, print_diagnostics,
update_diagnostics and stopping_metric.
Primal Dual Splitting Method (PDS)¶
Most algorithms shipped with Pycsou (FBS, APGD, ADMM, CPS, DRS, etc) are
special cases of Condat’s primal-dual splitting method (PDS)
implemented in the class PrimalDualSplitting (alias PDS):
class PrimalDualSplitting(dim: int,
F: Optional[DifferentiableMap] = None,
G: Optional[ProximableFunctional] = None,
H: Optional[ProximableFunctional] = None,
K: Optional[LinearOperator] = None,
tau: Optional[float] = None,
sigma: Optional[float] = None,
rho: Optional[float] = None,
beta: Optional[float] = None,
...)
The user must simply put its problem in the form \({\min_{\mathbf{x}\in\mathbb{R}^N} \;\mathcal{F}(\mathbf{x})\;\;+\;\;\mathcal{G}(\mathbf{x})\;\;+\;\;\mathcal{H}(\mathbf{K} \mathbf{x})}\) and provide the relevant terms to the class constructor.
See also
See Proximal Algorithms for implementations of the above mentionned algorithms in Pycsou.
Hyperparameters Auto-tuning¶
For convergence of the algorithm, the step sizes/momentum parameter \(\tau\), \(\sigma\) and \(\rho\) must verify:
If the Lipschitz constant \(\beta\) of \(\nabla \mathcal{F}\) is positive:
\(\frac{1}{\tau}-\sigma\Vert\mathbf{K}\Vert_{2}^2\geq \frac{\beta}{2}\),
\(\rho \in ]0,\delta[\), where \(\delta:=2-\frac{\beta}{2}\left(\frac{1}{\tau}-\sigma\Vert\mathbf{K}\Vert_{2}^2\right)^{-1}\in[1,2[.\)
If the Lipschitz constant \(\beta\) of \(\nabla \mathcal{F}\) is null (e.g. \(\mathcal{F}=0\)):
\(\tau\sigma\Vert\mathbf{K}\Vert_{2}^2\leq 1\)
\(\rho \in [\epsilon,2-\epsilon]\), for some \(\epsilon>0.\)
When the user does not specify hyperparameters, we choose the step sizes as large as possible and perfectly balanced (improves practical convergence speed):
\(\beta>0\):
\[\tau=\sigma=\frac{1}{\Vert\mathbf{K}\Vert_{2}^2}\left(-\frac{\beta}{4}+\sqrt{\frac{\beta^2}{16}+\Vert\mathbf{K}\Vert_{2}^2}\right).\]\(\beta=0\):
\[\tau=\sigma=\Vert\mathbf{K}\Vert_{2}^{-1}.\]
The momentum term \(\rho\) is chosen as \(\rho=0.9\) (\(\beta>0\)) or \(\rho=1\) (\(\beta=0\)).
Note
\(\Vert\mathbf{K}\Vert_{2}\) can be computed efficiently via the
method LinearOperator.compute_lipschitz_cst which relies on
Scipy’s sparse linear algebra routines eigs/eigsh/svds.
Example:¶
with \(\mathbf{D}\in\mathbb{R}^{N\times N}\) and \(\mathbf{G}\in\mathbb{R}^{L\times N}, \, \mathbf{y}\in\mathbb{R}^L, \lambda_1,\lambda_2>0.\) This problem can be written in the form
by choosing \(\mathcal{F}(\mathbf{x})= \frac{1}{2}\left\|\mathbf{y}-\mathbf{G}\mathbf{x}\right\|_2^2\), \(\mathcal{G}(\mathbf{x})=\lambda_2\|\mathbf{x}\|_1\), \(\mathcal{H}(\mathbf{x})=\lambda_1 \|\mathbf{x}\|_1\) and \(\mathbf{K}=\mathbf{D}\).
from pycsou.linop import FirstDerivative, DownSampling
from pycsou.func import SquaredL2Loss, L1Norm, NonNegativeOrthant
from pycsou.opt import PrimalDualSplitting
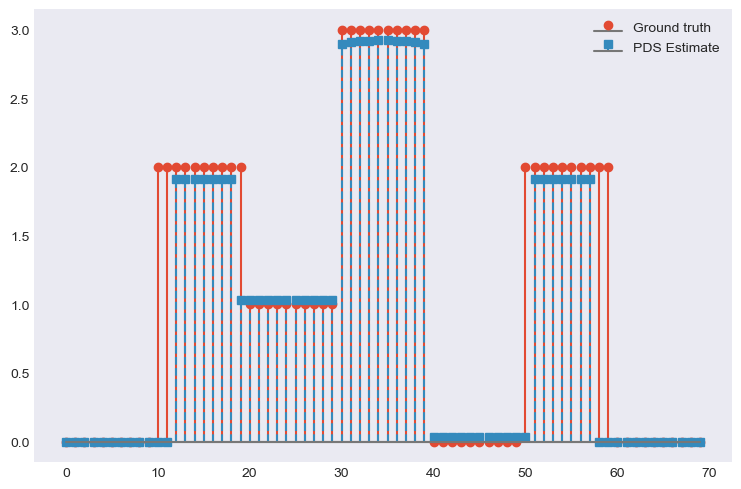
x = np.repeat([0, 2, 1, 3, 0, 2, 0], 10) # Ground truth
D = FirstDerivative(size=x.size, kind='forward') # Regularisation operator
D.compute_lipschitz_cst(tol=1e-3)
G = DownSampling(size=x.size, downsampling_factor=3) # Downsampling operator
G.compute_lipschitz_cst() # Compute Lipschitz constant for automatic parameter tuning
y = G(x) # Input data (downsampled x)
l22_loss = (1 / 2) * SquaredL2Loss(dim=G.shape[0], data=y) # Least-squares loss
F = l22_loss * G # Differentiable term F
H = 0.1 * L1Norm(dim=D.shape[0]) # Proximable term H
G = 0.01 * L1Norm(dim=G.shape[1]) # Proximable term F
pds = PrimalDualSplitting(dim=G.shape[1], F=F, G=G, H=H, K=D, verbose=None) # Initialise PDS
estimate, converged, diagnostics = pds.iterate() # Run PDS
plt.figure()
plt.stem(x, linefmt='C0-', markerfmt='C0o')
plt.stem(estimate['primal_variable'], linefmt='C1--', markerfmt='C1s')
plt.legend(['Ground truth', 'PDS Estimate'])
plt.show()