Sampling Operators¶
Module: pycsou.linop.sampling
Sampling operators.
This module provides sampling operators for discrete or continuous signals.
Discrete Sampling Operators
|
Subsampling operator. |
|
Masking operator. |
|
Downsampling operator. |
|
Pooling operator. |
Continuous-Domain Sampling Operators
|
Nearest neighbours sampling operator. |
|
Generalised Vandermonde matrix. |
|
Transformed Distance Matrix. |
-
SubSampling(size: int, sampling_indices: Union[numpy.ndarray, list], shape: Optional[tuple] = None, axis: int = 0, dtype: str = 'float64', inplace: bool = True)[source]¶ Subsampling operator.
Extract subset of values from input array at locations
sampling_indicesin forward mode and place those values at locationssampling_indicesin an otherwise zero array in adjoint mode.- Parameters
size (int) – Size of input array.
sampling_indices (
listornumpy.ndarray) – Integer indices of samples for data selection.shape (tuple) – Shape of input array (
Noneif only one dimension is available).axis (int) – When
shapeis notNone, axis along which subsampling is applied.dtype (str) – Type of elements in input array.
inplace (bool) – Work inplace (
True) or make a new copy (False). By default, data is a reference to the model (in forward) and model is a reference to the data (in adjoint).
- Returns
The subsampling operator.
- Return type
- Raises
ValueError – If shape and size do not match.
Examples
>>> x = np.arange(9).reshape(3,3) >>> sampling_indices = [0,2] >>> SamplingOp=SubSampling(size=x.size, sampling_indices=sampling_indices) >>> SamplingOp * x.reshape(-1) array([0, 2]) >>> SamplingOp.adjoint(SamplingOp* x.reshape(-1)).reshape(x.shape) array([[0., 0., 2.], [0., 0., 0.], [0., 0., 0.]]) >>> SamplingOp=SubSampling(size=x.size, sampling_indices=sampling_indices, shape=x.shape, axis=1) >>> (SamplingOp * x.reshape(-1)).reshape(x.shape[1], len(sampling_indices)) array([[0, 2], [3, 5], [6, 8]]) >>> SamplingOp.adjoint(SamplingOp* x.reshape(-1)).reshape(x.shape) array([[0., 0., 2.], [3., 0., 5.], [6., 0., 8.]])
Notes
Subsampling of a subset of \(L\) values at locations
sampling_indicesfrom an input vector \(\mathbf{x}\) of size \(N\) can be expressed as:\[y_i = x_{n_i} \quad \forall i=1,2,...,L,\]where \(\mathbf{n}=[n_1, n_2,..., n_L]\) is a vector containing the indeces of the original array at which samples are taken.
Conversely, in adjoint mode the available values in the data vector \(\mathbf{y}\) are placed at locations \(\mathbf{n}=[n_1, n_2,..., n_L]\) in the model vector:
\[x_{n_i} = y_i \quad \forall i=1,2,...,L\]and \(x_{j}=0 \,\forall j \neq n_i\) (i.e., at all other locations in input vector).
See also
Masking,Downsampling
-
class
Masking(size: int, sampling_bool: Union[numpy.ndarray, list], dtype: type = <class 'numpy.float64'>)[source]¶ Bases:
pycsou.core.linop.LinearOperatorMasking operator.
Extract subset of values from input array at locations marked as
Trueinsampling_bool.Examples
>>> x = np.arange(9).reshape(3,3) >>> sampling_bool = np.zeros((3,3)).astype(bool) >>> sampling_bool[[1,2],[0,2]] = True >>> SamplingOp = Masking(size=x.size, sampling_bool=sampling_bool) >>> SamplingOp * x.reshape(-1) array([3, 8]) >>> SamplingOp.adjoint(SamplingOp* x.reshape(-1)).reshape(x.shape) array([[0., 0., 0.], [3., 0., 0.], [0., 0., 8.]]) >>> sampling_indices = np.nonzero(sampling_bool.reshape(-1))[0].astype(int) >>> SubSamplingOp=SubSampling(size=x.size, sampling_indices=sampling_indices) >>> np.allclose(SamplingOp * x.reshape(-1), SubSamplingOp * x.reshape(-1)) True
Notes
For flattened arrays, the
Maskingoperator is equivalent to theSubSampling()operator, with the only difference that the sampling locations are specified in the form of a boolean array instead of indices.See also
SubSampling,Downsampling-
__init__(size: int, sampling_bool: Union[numpy.ndarray, list], dtype: type = <class 'numpy.float64'>)[source]¶ - Parameters
size (int) – Size of input array.
sampling_bool (
listornumpy.ndarray) – Boolean array for data selection.Truevalues mark the positions of the samples.dtype (str) –
of elements in input array. (Type) –
- Raises
ValueError – If the size of
sampling_booldiffers fromsize.
-
-
class
DownSampling(size: int, downsampling_factor: Union[int, tuple, list], shape: Optional[tuple] = None, axis: Optional[int] = None, dtype: type = <class 'numpy.float64'>)[source]¶ Bases:
pycsou.linop.sampling.MaskingDownsampling operator.
Downsample an array in one of the three ways:
If
shapeisNone: The input array is flat and the downsampling selects one element everydownsampling_factorelements.If
shapeis notNoneandaxisisNone: The input array is multidimensional, and each dimension is downsampled by a certain factor. Downsampling factors for each dimension are specified in the tupledownsampling_factor(ifdownsampling_factoris an integer, then the same factor is assumed for every dimension).If
shapeis notNoneandaxisis notNone: The input array is multidimensional, but only the dimension is specified byaxisis downsampled by adownsampling_factor.
Examples
>>> x = np.arange(10) >>> DownSamplingOp = DownSampling(size=x.size, downsampling_factor=3) >>> DownSamplingOp * x array([0, 3, 6, 9]) >>> DownSamplingOp.adjoint(DownSamplingOp * x) array([0., 0., 0., 3., 0., 0., 6., 0., 0., 9.]) >>> x = np.arange(20).reshape(4,5) >>> DownSamplingOp = DownSampling(size=x.size, shape=x.shape, downsampling_factor=3) >>> (DownSamplingOp * x.flatten()).reshape(DownSamplingOp.output_shape) array([[ 0, 3], [15, 18]]) >>> DownSamplingOp = DownSampling(size=x.size, shape=x.shape, downsampling_factor=(2,4)) >>> (DownSamplingOp * x.flatten()).reshape(DownSamplingOp.output_shape) array([[ 0, 4], [10, 14]]) >>> DownSamplingOp.adjoint(DownSamplingOp * x.flatten()).reshape(x.shape) array([[ 0., 0., 0., 0., 4.], [ 0., 0., 0., 0., 0.], [10., 0., 0., 0., 14.], [ 0., 0., 0., 0., 0.]]) >>> DownSamplingOp = DownSampling(size=x.size, shape=x.shape, downsampling_factor=2, axis=-1) >>> (DownSamplingOp * x.flatten()).reshape(DownSamplingOp.output_shape) array([[ 0, 2, 4], [ 5, 7, 9], [10, 12, 14], [15, 17, 19]]) >>> DownSamplingOp.adjoint(DownSamplingOp * x.flatten()).reshape(x.shape) array([[ 0., 0., 2., 0., 4.], [ 5., 0., 7., 0., 9.], [10., 0., 12., 0., 14.], [15., 0., 17., 0., 19.]])
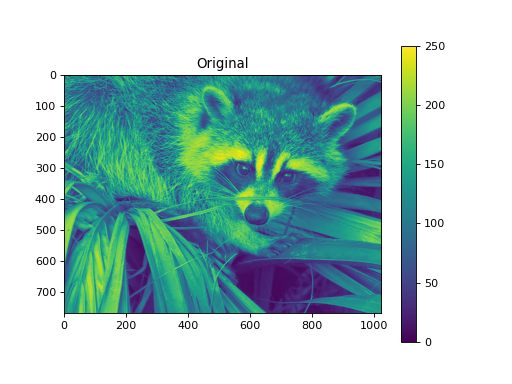
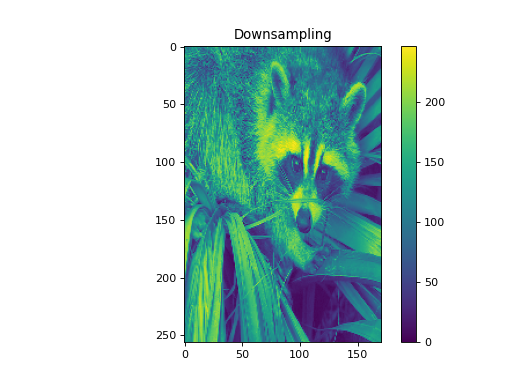
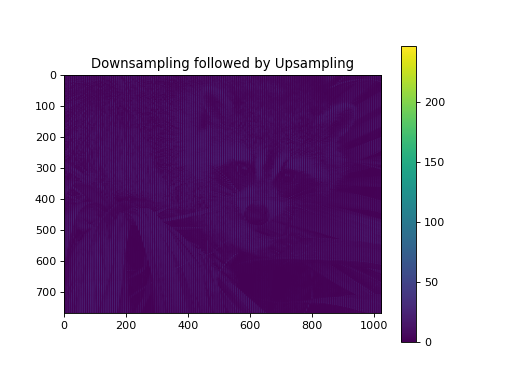
import numpy as np from pycsou.linop.sampling import DownSampling import matplotlib.pyplot as plt import scipy.misc img = scipy.misc.face(gray=True).astype(float) DownSampOp = DownSampling(size=img.size, shape=img.shape, downsampling_factor=(3,6)) down_sampled_img = (DownSampOp * img.flatten()).reshape(DownSampOp.output_shape) up_sampled_img = DownSampOp.adjoint(down_sampled_img.flatten()).reshape(img.shape) plt.figure() plt.imshow(img) plt.colorbar() plt.title('Original') plt.figure() plt.imshow(down_sampled_img) plt.colorbar() plt.title('Downsampling') plt.figure() plt.imshow(up_sampled_img) plt.colorbar() plt.title('Downsampling followed by Upsampling')
Notes
Downsampling by \(M\) an input vector \(\mathbf{x}\) of size \(N\) can be performed as:
\[y_i = x_{iM} \quad i=1,\ldots, \lfloor N/M \rfloor.\]Conversely, in adjoint mode the available values in the data vector \(\mathbf{y}\) are placed at locations \(n=iM\) in the model vector:
\[x_{iM} = y_i,\;\;x_{iM+1}=x_{iM+2}=\cdots=x_{(i+1)M-1}=0, \qquad i=1,\ldots, \lfloor N/M \rfloor.\]See also
-
__init__(size: int, downsampling_factor: Union[int, tuple, list], shape: Optional[tuple] = None, axis: Optional[int] = None, dtype: type = <class 'numpy.float64'>)[source]¶ - Parameters
size (int) – Size of input array.
downsampling_factor (Union[int, tuple, list]) – Downsampling factor (possibly different for each dimension).
shape (Optional[tuple]) – Shape of input array (default
None: the input array is 1D).axis (Optional[int]) – Axis along which to downsample for ND input arrays (default
None: downsampling is performed along each axis).dtype (type) – Type of input array.
- Raises
ValueError – If the set of parameters {
shape,size,sampling_factor,axis} is invalid.
-
class
Pooling(shape: tuple, block_size: Union[tuple, list], pooling_func: str = 'mean', dtype: type = <class 'numpy.float64'>)[source]¶ Bases:
pycsou.core.linop.LinearOperatorPooling operator.
Pool an array by summing/averaging across constant size blocks tiling the array.
Examples
>>> x = np.arange(24).reshape(4,6) >>> PoolingOp = Pooling(shape=x.shape, block_size=(2,3), pooling_func='mean') >>> (PoolingOp * x.flatten()).reshape(PoolingOp.output_shape) array([[ 4., 7.], [16., 19.]]) >>> PoolingOp.adjoint(PoolingOp * x.flatten()).reshape(x.shape) array([[ 4., 4., 4., 7., 7., 7.], [ 4., 4., 4., 7., 7., 7.], [16., 16., 16., 19., 19., 19.], [16., 16., 16., 19., 19., 19.]]) >>> PoolingOp = Pooling(shape=x.shape, block_size=(2,3), pooling_func='sum') >>> (PoolingOp * x.flatten()).reshape(PoolingOp.output_shape) array([[ 24, 42], [ 96, 114]]) >>> PoolingOp.adjoint(PoolingOp * x.flatten()).reshape(x.shape) array([[ 24, 24, 24, 42, 42, 42], [ 24, 24, 24, 42, 42, 42], [ 96, 96, 96, 114, 114, 114], [ 96, 96, 96, 114, 114, 114]])
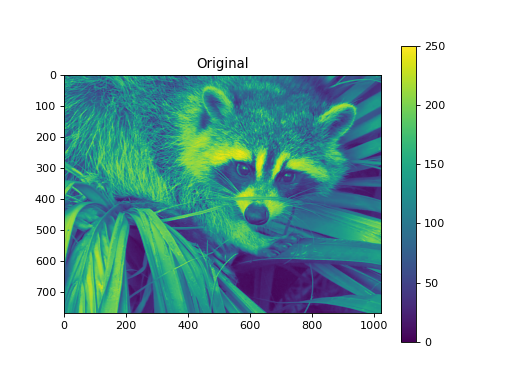
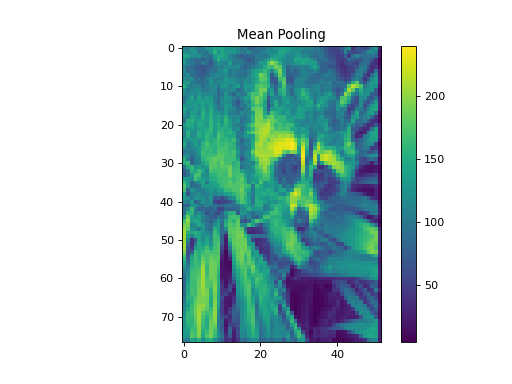
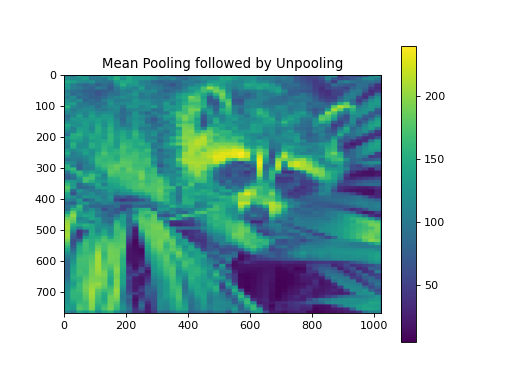
import numpy as np from pycsou.linop.sampling import Pooling import matplotlib.pyplot as plt import scipy.misc img = scipy.misc.face(gray=True).astype(float) PoolingOp = Pooling(shape=img.shape, block_size=(10,20)) pooled_img = (PoolingOp * img.flatten()).reshape(PoolingOp.output_shape) adjoint_img = PoolingOp.adjoint(pooled_img.flatten()).reshape(img.shape) plt.figure() plt.imshow(img) plt.colorbar() plt.title('Original') plt.figure() plt.imshow(pooled_img) plt.colorbar() plt.title('Mean Pooling') plt.figure() plt.imshow(adjoint_img) plt.colorbar() plt.title('Mean Pooling followed by Unpooling')
Notes
Pooling is performed via the function skimage.measure.block_reduce from
scikit-image. If one dimension of the image is not perfectly divisible by the block size then it is zero padded.The adjoint (unpooling) is performed by assigning the value of the blocks through the pooling function (e.g. mean, sum) to each element of the blocks.
Warning
Max, median or min pooling are not supported since the resulting PoolingOperator would then be non linear!
See also
SubSampling,Downsampling-
__init__(shape: tuple, block_size: Union[tuple, list], pooling_func: str = 'mean', dtype: type = <class 'numpy.float64'>)[source]¶ - Parameters
- Raises
ValueError – If the block size is inconsistent with the input array shape or if the pooling function is not supported.
-
-
class
NNSampling(samples: numpy.ndarray, grid: numpy.ndarray, dtype: type = <class 'numpy.float64'>)[source]¶ Bases:
pycsou.core.linop.LinearOperatorNearest neighbours sampling operator.
Sample a gridded ND signal at on-grid nearest neighbours of off-grid sampling locations. This can be useful when piecewise constant priors are used to recover continuously-defined signals sampled non-uniformly (see [FuncSphere] Remark 6.9).
Examples
>>> rng = np.random.default_rng(seed=0) >>> x = np.arange(24).reshape(4,6) >>> grid = np.stack(np.meshgrid(np.arange(6),np.arange(4)), axis=-1) >>> samples = np.stack((5 * rng.random(size=6),3 * rng.random(size=6)), axis=-1) >>> print(samples) [[3.18480844 1.81990733] [1.34893357 2.18848968] [0.20486762 1.63087497] [0.08263818 2.80521727] [4.0663512 2.44756066] [4.56377789 0.0082155 ]] >>> NNSamplingOp = NNSampling(samples=samples, grid=grid) >>> (NNSamplingOp * x.flatten()) array([15, 13, 12, 18, 16, 5]) >>> NNSamplingOp.adjoint(NNSamplingOp * x.flatten()).reshape(x.shape) array([[ 0., 0., 0., 0., 0., 5.], [ 0., 0., 0., 0., 0., 0.], [12., 13., 0., 15., 16., 0.], [18., 0., 0., 0., 0., 0.]])
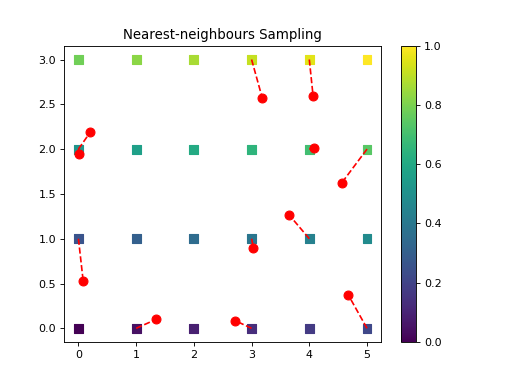
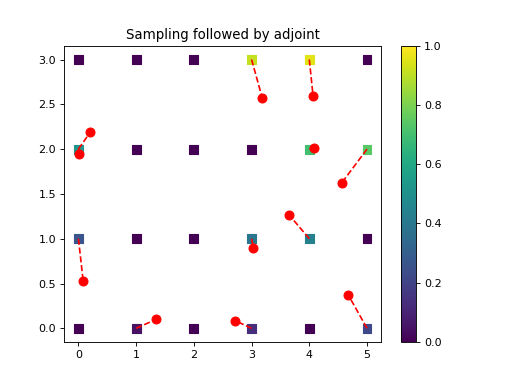
import numpy as np from pycsou.linop.sampling import NNSampling import matplotlib.pyplot as plt rng = np.random.default_rng(seed=0) rng = np.random.default_rng(seed=0) x = np.arange(24).reshape(4, 6) grid = np.stack(np.meshgrid(np.arange(6), np.arange(4)), axis=-1) samples = np.stack((5 * rng.random(size=12), 3 * rng.random(size=12)), axis=-1) NNSamplingOp = NNSampling(samples=samples, grid=grid) grid = grid.reshape(-1, 2) x = x.reshape(-1) y = (NNSamplingOp * x.flatten()) x_samp = NNSamplingOp.adjoint(y).reshape(x.shape) plt.scatter(grid[..., 0].reshape(-1), grid[..., 1].reshape(-1), s=64, c=x.reshape(-1), marker='s', vmin=np.min(x), vmax=np.max(x)) plt.scatter(samples[:, 0], samples[:, 1], c='r', s=64) plt.plot(np.stack((grid[NNSamplingOp.nn_indices, 0], samples[:, 0]), axis=0), np.stack((grid[NNSamplingOp.nn_indices, 1], samples[:, 1]), axis=0), '--r') plt.colorbar() plt.title('Nearest-neighbours Sampling') plt.figure() plt.scatter(grid[..., 0].reshape(-1), grid[..., 1].reshape(-1), s=64, c=x_samp.reshape(-1), marker='s', vmin=np.min(x), vmax=np.max(x)) plt.scatter(samples[:, 0], samples[:, 1], c='r', s=64) plt.plot(np.stack((grid[NNSamplingOp.nn_indices, 0], samples[:, 0]), axis=0), np.stack((grid[NNSamplingOp.nn_indices, 1], samples[:, 1]), axis=0), '--r') plt.colorbar() plt.title('Sampling followed by adjoint')
Notes
Consider a signal defined over a mesh \(f:\{\mathbf{n}_1,\ldots, \mathbf{n}_M\}\to \mathbb{C}\), with \(\{\mathbf{n}_1,\ldots, \mathbf{n}_M\}\subset \mathbb{R}^N\). Consider moreover sampling locations \(\{\mathbf{z}_1,\ldots, \mathbf{z}_L\}\subset \mathbb{R}^N\) which do not necessarily lie on the mesh. Then, nearest-neighbours sampling is defined as:
\[y_i=f\left[\arg\min_{k=1,\ldots, M}\|\mathbf{z}_i-\mathbf{n}_k\|_2\right], \qquad i=1,\ldots, L.\]Note that in practice every sample locations has exactly one nearest neighbour (ties have probability zero) and hence this equation is well-defined.
Given a vector \(\mathbf{y}=[y_1,\ldots, y_L]\in\mathbb{C}^N\), the adjoint of the
NNSamplingoperator is defined as\[f[\mathbf{n}_k]=\mbox{mean}\left\{y_i :\; \mathbf{n}_k=\arg\min_{j=1,\ldots,M}\|\mathbf{z}_i-\mathbf{n}_j\|_2,\, i=1,\ldots, L\right\},\quad k=1,\ldots, M,\]where \(\mbox{mean}\{B\}=|B|^{-1}\sum_{z\in B} z,\; \forall B\subset\mathbb{C}\) and with the convention that \(\mbox{mean}\{\emptyset\}=0.\) The mean is used to handle cases where many sampling locations are mapped to a common nearest neighbour on the mesh.
Warning
The grid needs not be uniform! Think of it as a mesh. It can happen that more than one sampling location is mapped to the same nearest neighbour.
See also
-
__init__(samples: numpy.ndarray, grid: numpy.ndarray, dtype: type = <class 'numpy.float64'>)[source]¶ - Parameters
samples (np.ndarray) – Off-grid sampling locations with shape (M,N).
grid – Grid points with shape (L,N).
dtype (type) – Type of the input array.
- Raises
ValueError – If the dimension of the sample locations and the grid points do not match.
-
-
class
GeneralisedVandermonde(samples: numpy.ndarray, funcs: List[Callable], dtype: type = <class 'numpy.float64'>)[source]¶ Bases:
pycsou.linop.base.DenseLinearOperatorGeneralised Vandermonde matrix.
Given sampling locations \(\{\mathbf{z}_1,\ldots,\mathbf{z}_L\}\subset\mathbb{R}^N\), and a family of continuous functions \(\{\varphi_1, \ldots, \varphi_K\}\subset \mathcal{C}(\mathbb{R}^N, \mathbb{C})\), this function forms the generalised Vandermonde matrix:
\[\begin{split}\left[\begin{array}{ccc} \varphi_1(\mathbf{z}_1) & \cdots & \varphi_K(\mathbf{z}_1)\\\vdots & \ddots & \vdots\\\varphi_1(\mathbf{z}_L) & \cdots & \varphi_K(\mathbf{z}_L)\end{array}\right]\in\mathbb{C}^{L\times K}.\end{split}\]This matrix is useful for sampling functions in the span of \(\{\varphi_1, \ldots, \varphi_K\}\). Indeed, if \(f=\sum_{k=1}^K\alpha_k\varphi_k\), then we have
\[\begin{split}\left[\begin{array}{c}f(\mathbf{z}_1)\\\vdots\\ f(\mathbf{z}_L) \end{array}\right]=\left[\begin{array}{ccc} \varphi_1(\mathbf{z}_1) & \cdots & \varphi_K(\mathbf{z}_1)\\\vdots & \ddots & \vdots\\\varphi_1(\mathbf{z}_L) & \cdots & \varphi_K(\mathbf{z}_L)\end{array}\right]\left[\begin{array}{c}\alpha_1\\\vdots\\ \alpha_K \end{array}\right].\end{split}\]Examples
>>> samples = np.arange(10) >>> func1 = lambda t: t**2; func2 = lambda t: t**3; funcs = [func1, func2] >>> VOp = GeneralisedVandermonde(funcs=funcs, samples=samples) >>> alpha=np.ones((2,)) >>> VOp.mat array([[ 0., 0.], [ 1., 1.], [ 4., 8.], [ 9., 27.], [ 16., 64.], [ 25., 125.], [ 36., 216.], [ 49., 343.], [ 64., 512.], [ 81., 729.]]) >>> np.allclose(VOp * alpha, samples ** 2 + samples ** 3) True
See also
-
class
MappedDistanceMatrix(samples1: numpy.ndarray, function: Callable, samples2: Optional[numpy.ndarray] = None, mode: str = 'radial', max_distance: Optional[float] = None, dtype: type = <class 'numpy.float64'>, chunks: Union[str, int, tuple, None] = 'auto', operator_type: str = 'dask', verbose: bool = True, n_jobs: int = -1, joblib_backend: str = 'loky', ord: float = 2.0, eps: float = 0)[source]¶ Bases:
pycsou.linop.base.ExplicitLinearOperatorTransformed Distance Matrix.
Given two point sets \(\{\mathbf{z}_1,\ldots,\mathbf{z}_L\}\subset\mathbb{R}^N\), \(\{\mathbf{x}_1,\ldots,\mathbf{x}_K\}\subset\mathbb{R}^N\) and a continuous function \(\varphi:\mathbb{R}_+\to\mathbb{C}\), this function forms the following matrix:
\[\begin{split}\left[\begin{array}{ccc} \varphi(d(\mathbf{z}_1,\mathbf{x}_1)) & \cdots & \varphi(d(\mathbf{z}_1,\mathbf{x}_K))\\\vdots & \ddots & \vdots\\\varphi(d(\mathbf{z}_L,\mathbf{x}_1)) & \cdots & \varphi(d(\mathbf{z}_1,\mathbf{x}_K))\end{array}\right]\in\mathbb{C}^{L\times K},\end{split}\]where \(d:\mathbb{R}^N\times \mathbb{R}^N\to \mathbb{R}\) is a (signed) distance defined in one of the following two ways:
Radial: \(d(\mathbf{z},\mathbf{x})=\|\mathbf{z}-\mathbf{x}\|_p, \; \forall \mathbf{z},\mathbf{x}\in\mathbb{R}^N,\) with \(p\in [1, +\infty]\).
Zonal: \(d(\mathbf{z},\mathbf{x})=\langle\mathbf{z},\mathbf{x}\rangle, \; \forall \mathbf{z},\mathbf{x}\in\mathbb{S}^{N-1}.\) Note that in this case the two point sets must be on the hypersphere \(\mathbb{S}^{N-1}.\)
This matrix is useful for sampling sums of radial/zonal functions. Indeed, if \(f(\mathbf{z})=\sum_{k=1}^K\alpha_k\varphi(d(\mathbf{z},\mathbf{x}_k))\), then we have
\[\begin{split}\left[\begin{array}{c}f(\mathbf{z}_1)\\\vdots\\ f(\mathbf{z}_L) \end{array}\right]=\left[\begin{array}{ccc} \varphi(d(\mathbf{z}_1,\mathbf{x}_1)) & \cdots & \varphi(d(\mathbf{z}_1,\mathbf{x}_K))\\\vdots & \ddots & \vdots\\\varphi(d(\mathbf{z}_L,\mathbf{x}_1)) & \cdots & \varphi(d(\mathbf{z}_1,\mathbf{x}_K))\end{array}\right]\left[\begin{array}{c}\alpha_1\\\vdots\\ \alpha_K \end{array}\right].\end{split}\]Examples
>>> rng = np.random.default_rng(seed=1) >>> sigma = 1 / 12 >>> func = lambda x: np.exp(-x ** 2 / (2 * sigma ** 2)) >>> max_distance = 3 * sigma >>> samples1 = np.linspace(0, 2, 10); samples2 = rng.random(size=3) >>> MDMOp1 = MappedDistanceMatrix(samples1=samples1, samples2=samples2, function=func,max_distance=max_distance, operator_type='sparse', n_jobs=-1) >>> MDMOp2 = MappedDistanceMatrix(samples1=samples1, samples2=None, function=func,max_distance=max_distance, operator_type='sparse', n_jobs=-1) >>> MDMOp3 = MappedDistanceMatrix(samples1=samples1, samples2=samples2, function=func, operator_type='dense') >>> MDMOp4 = MappedDistanceMatrix(samples1=samples1, samples2=samples2, function=func, operator_type='dask') >>> type(MDMOp1.mat), type(MDMOp3.mat), type(MDMOp4.mat) (<class 'scipy.sparse.csr.csr_matrix'>, <class 'numpy.ndarray'>, <class 'dask.array.core.Array'>) >>> MDMOp1.mat.shape, MDMOp2.mat.shape ((10, 3), (10, 10)) >>> MDMOp3.mat array([[6.43689834e-009, 5.64922608e-029, 2.23956473e-001], [2.38517543e-003, 2.61112241e-017, 6.44840995e-001], [7.21186665e-001, 9.84802612e-009, 1.51504434e-003], [1.77933914e-001, 3.03078296e-003, 2.90456923e-009], [3.58222996e-005, 7.61104282e-001, 4.54382719e-018], [5.88480228e-012, 1.55961419e-001, 5.80023464e-030], [7.88849171e-022, 2.60779757e-005, 6.04161441e-045], [8.62858845e-035, 3.55806811e-012, 5.13504354e-063], [7.70139186e-051, 3.96130538e-022, 3.56138512e-084], [5.60896038e-070, 3.59870356e-035, 2.01547509e-108]]) >>> alpha = rng.lognormal(size=samples2.size, sigma=0.5) >>> beta = rng.lognormal(size=samples1.size, sigma=0.5) >>> MDMOp1 * alpha array([0.27995783, 0.8060865 , 0.37589858, 0.09274313, 1.19684992, 0.24525208, 0. , 0. , 0. , 0. ]) >>> MDMOp2 * beta array([0.80274037, 1.39329178, 1.27124579, 1.22168244, 1.08494933, 1.36310926, 0.7558366 , 0.96398702, 0.85066284, 1.37152693]) >>> MDMOp3 * alpha array([2.79957838e-01, 8.07329704e-01, 3.77792488e-01, 9.75090904e-02, 1.19686859e+00, 2.45252084e-01, 4.10080770e-05, 5.59512490e-12, 6.22922262e-22, 5.65902486e-35]) >>> MDMOp4 * alpha array([2.79957838e-01, 8.07329704e-01, 3.77792488e-01, 9.75090904e-02, 1.19686859e+00, 2.45252084e-01, 4.10080770e-05, 5.59512490e-12, 6.22922262e-22, 5.65902486e-35]) >>> np.allclose(MDMOp3 * alpha, MDMOp4 * alpha) True
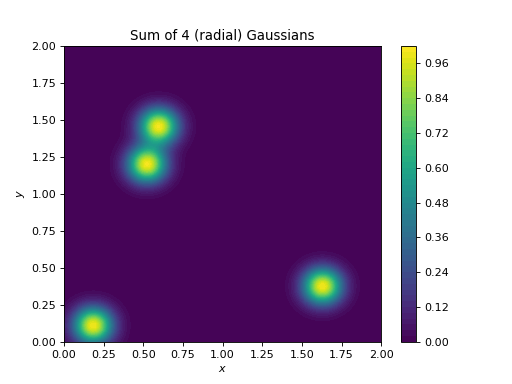
import numpy as np import matplotlib.pyplot as plt from pycsou.linop.sampling import MappedDistanceMatrix t = np.linspace(0, 2, 256) rng = np.random.default_rng(seed=2) x,y = np.meshgrid(t,t) samples1 = np.stack((x.flatten(), y.flatten()), axis=-1) samples2 = np.stack((2 * rng.random(size=4), 2 * rng.random(size=4)), axis=-1) alpha = np.ones(samples2.shape[0]) sigma = 1 / 12 func = lambda x: np.exp(-x ** 2 / (2 * sigma ** 2)) MDMOp = MappedDistanceMatrix(samples1=samples1, samples2=samples2, function=func, operator_type='dask') plt.contourf(x,y,(MDMOp * alpha).reshape(t.size, t.size), 50) plt.title('Sum of 4 (radial) Gaussians') plt.colorbar() plt.xlabel('$x$') plt.ylabel('$y$')
(Source code, png, hires.png, pdf)
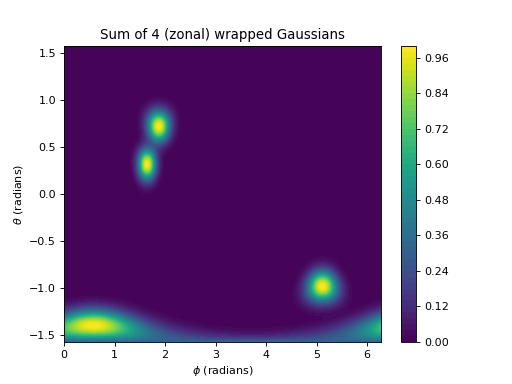
import numpy as np import matplotlib.pyplot as plt from pycsou.linop.sampling import MappedDistanceMatrix rng = np.random.default_rng(seed=2) phi = np.linspace(0,2*np.pi, 256) theta = np.linspace(-np.pi/2,np.pi/2, 256) phi, theta = np.meshgrid(phi, theta) x,y,z = np.cos(phi)*np.cos(theta), np.sin(phi)*np.cos(theta), np.sin(theta) samples1 = np.stack((x.flatten(), y.flatten(), z.flatten()), axis=-1) phi2 = 2 * np.pi * rng.random(size=4) theta2 = np.pi * rng.random(size=4) - np.pi/2 x2,y2,z2 = np.cos(phi2)*np.cos(theta2), np.sin(phi2)*np.cos(theta2), np.sin(theta2) samples2 = np.stack((x2,y2,z2), axis=-1) alpha = np.ones(samples2.shape[0]) sigma = 1 / 9 func = lambda x: np.exp(-np.abs(1-x) / (sigma ** 2)) MDMOp = MappedDistanceMatrix(samples1=samples1, samples2=samples2, function=func,mode='zonal', operator_type='sparse', max_distance=3*sigma) plt.contourf(phi, theta, (MDMOp * alpha).reshape(phi.shape), 50) plt.title('Sum of 4 (zonal) wrapped Gaussians') plt.colorbar() plt.xlabel('$\\phi$ (radians)') plt.ylabel('$\\theta$ (radians)')
(Source code, png, hires.png, pdf)
See also
-
__init__(samples1: numpy.ndarray, function: Callable, samples2: Optional[numpy.ndarray] = None, mode: str = 'radial', max_distance: Optional[float] = None, dtype: type = <class 'numpy.float64'>, chunks: Union[str, int, tuple, None] = 'auto', operator_type: str = 'dask', verbose: bool = True, n_jobs: int = -1, joblib_backend: str = 'loky', ord: float = 2.0, eps: float = 0)[source]¶ - Parameters
samples1 (np.ndarray) – First point set with shape (L,N).
function (Callable) – Function \(\varphi: \mathbb{R}_+\to \mathbb{C}\) to apply to each entry of the distance matrix.
samples2 (Optional[np.ndarray]) – Optional second point set with shape (K,N). If
None,samples2is equal tosamples1and the operator symmetric.mode (str) – How to compute the distances. If
'radial', the Euclidean distance with respect to the Minkowski norm \(\|\cdot\|_p\) is used. \(p\) is specified via the parameterordand has to meet the condition 1 <= p <= infinity. If'zonal'the spherical geodesic distance is used.max_distance (Optional[np.float]) – Support of the function \(\varphi\). Must be specified for sparse representation.
dtype (type) – Type of input array.
chunks (Union[str, int, tuple, None]) – Chunk sizes for Dask representation.
operator_type (str) – Internal representation of the operator:
'dense'represents the operator as a Numpy array,'dask'as a Dask array,'sparse'as a sparse matrix. Dask arrays or sparse matrices can be more memory efficient for very large point sets.verbose (bool) – Verbosity for parallel computations (only for sparse representations).
n_jobs (int) – Number of cores for parallel computations (only for sparse representations).
n_jobs=-1uses all available cores.joblib_backend (str) – Joblib backend (more details here).
ord (float) – Order of the norm for
mode='radial'.ordmust satisfy 1<= ord <=infty.eps (float) – Parameter for approximate nearest neighbours search when
mode='sparse'.
- Raises
ValueError – If
modeis invalid or ifoperator_typeis'sparse'butmax_distance=None.
-
get_sparse_mdm() → scipy.sparse.csr.csr_matrix[source]¶ Form the Mapped Distance Matrix as a sparse matrix.
- Returns
Sparse Mapped Distance Matrix.
- Return type
sparse.csr_matrix